RNA Chaperoning
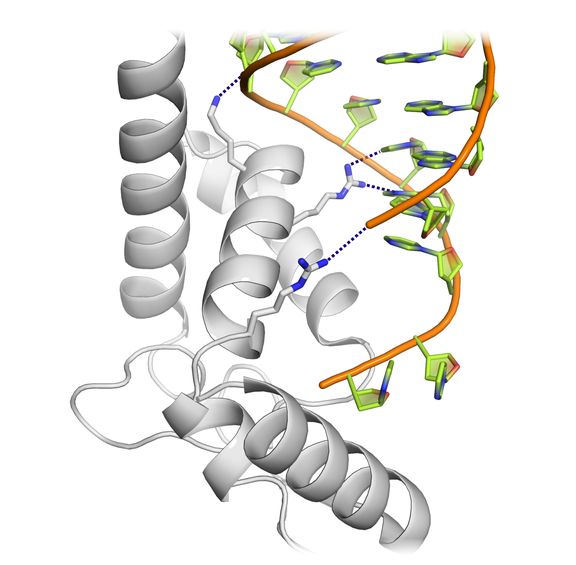
High-resolution structural information for RNA-protein binding is obtained through X-ray crystallography, using minimal interaction regions.
RNA chaperones play key regulating roles, e.g. in the control of gene expression.
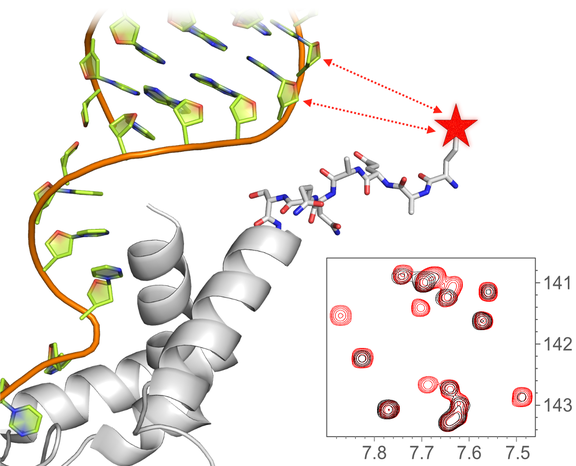
Long-range structural information is derived from paramagnetic relaxation NMR data, using PRE spin labels attached to the protein or the RNA.
