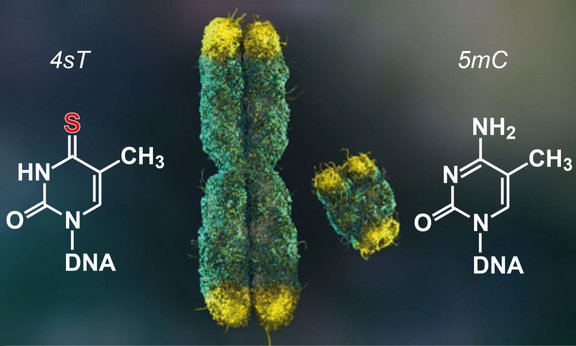
Chromosomes are the carriers of genetic information. In multiplying cell populations, chromosomes are constantly reorganized to ensure two fundamental functions: During cell division, a copy of the genetic information is mechanically transported to the daughter cells, while the genetic information is doubled and read between two cell divisions. In the study now published in Nature, researchers from the Vienna BioCenter and the Department of Organic Chemistry of the University of Innsbruck developed a new method that can map the relative spatial arrangement of the two replicated sister DNA molecules in each chromosome.
Genomic map
The method is based on a chemical DNA labeling method and high-throughput sequence analysis, which generates a genomic map of contact points between labeled and unlabeled sister DNA molecules. With this new method, the function of key factors of the chromosome organization, such as cohesin, has been be elucidated. The new technology of 'Sister chromatid-sensitive chromosome conformation capture (scsHi-C)' will make it possible to address a broad range of fundamental biological questions in the future, e.g. the organization of the genome in DNA repair processes or in recombination events of meiosis. The work thus provides the basis for gaining important knowledge about cellular genome inheritance and establishes an innovative technology for the investigation of DNA folding structures.
Chemistry to success
The chemical key to success of the study lies in the osmium tetroxide-based conversion of thionucleosides. This chemistry developed in the group of Ronald Micura was also the breakthrough for a previously published novel RNA sequencing approach (TUC-seq) allowing mRNA life cycle determinations and now has been expanded successfully to DNA applications.
Links
- Conformation of sister chromatids in the replicated human genome. Michael Mitter, Catherina Gasser, Zsuzsanna Takacs, Christoph C. H. Langer, Wen Tang, Gregor Jessberger, Charlie T. Beales, Eva Neuner, Stefan L. Ameres, Jan-Michael Peters, Anton Goloborodko, Ronald Micura, Daniel W. Gerlich, Nature 2020
- Forschungsgruppe Ronald Micura
- Forschungsgruppe Daniel Gerlich
- Forschungsgruppe Daniel Gerlich
- RNA dynamics revealed by metabolic RNA labeling and biochemical nucleoside conversions. MAP Baptista, Lars Dölken, Nature Methods 2018, 15, 171–172.
- Centrum für Molekulare Biowissenschaften (CMBI), Universität Innsbruck